Documentation How to Use SchizConnect
We hope that this documentation will help answer any questions you may have about using SchizConnect and its data. If you aren't finding what you need, please let us know. Please also check out our documentation by project.
Frequently Asked Questions
Tell me about the projects in SchizConnect.
SchizConnect currently contains data from the following projects:
- BrainGluSchi (COINS): This is a case control study to examine glutamate+glutamine as well as other traditional neurometabolites in a large sample of schizophrenia and healthy control subjects over a broad age range.
- COBRE (COINS): The Center for Biomedical Research Excellence (COBRE) examines the neural mechanisms of schizophrenia by integrating multiple neuroimaging methods with psychiatric, neuropsychological and genetic testing.
- fBIRNPhaseII__0010 (UCI_HID): Chronic schizophrenic patients and age-matched controls were recruited from multiple sites and all performed the same diagnostic and symptom assessments, and the same fMRI study. Each subject was scanned twice within a one-month period if at all possible. The fMRI study consisted of a sensorimotor scan, an auditory oddball task, a working memory task, and a breath-holding task. Patients and controls are matched by gender, handedness and age within 5 years. Schizophrenics have more mixed hand dominance than controls. In such cases they usually are not truly ambidextrous, but have less clear dominance. In such cases, we will go with the most prominent dominance for purposes of classification. We have not chosen to match on race.
- MCICShare (COINS): The MIND Clinical Imaging Consortium (MCIC) contains comprehensive clinical data, and raw structural, functional and diffusion-weighted DICOM images in schizophrenia patients and sex and age-matched controls.
- NMorphCH (NU_REDCAP, NUNDA): This is a longitudinal study examining the clinical, cognitive and neuroimaging (MRI) data from schizophrenia and control subjects at baseline and after two years. The study was conducted at Northwestern University. Neuroimaging data includes T1, T2, DTI, resting-state fMRI and n-back task fMRI. Clinical data includes symptom and mood assessments. Cognitive data includes neuropsychological assessments on working memory, episodic memory, and executive function domains.
- NUSDAST (XNAT_CENTRAL, NU_REDCAP): The Northwestern University Schizophrenia Data and Software Tool (NUSDAST) is a repository of schizophrenia neuroimaging data collected from over 450 individuals with schizophrenia, healthy controls and their respective siblings, most with 2-year (some with 4-year) longitudinal follow-up.
- REWARD (COINS): The goal of the REWARD project was to use state-of-the art functional neuroimaging and behavioral paradigms derived from the affective science literature to examine the integrity of the components of the neural systems linking rewards and actions among individuals with schizophrenia, health controls and the siblings of individuals with schizophrenia.
How do I download ALL images or assessments for the subjects my query returns?
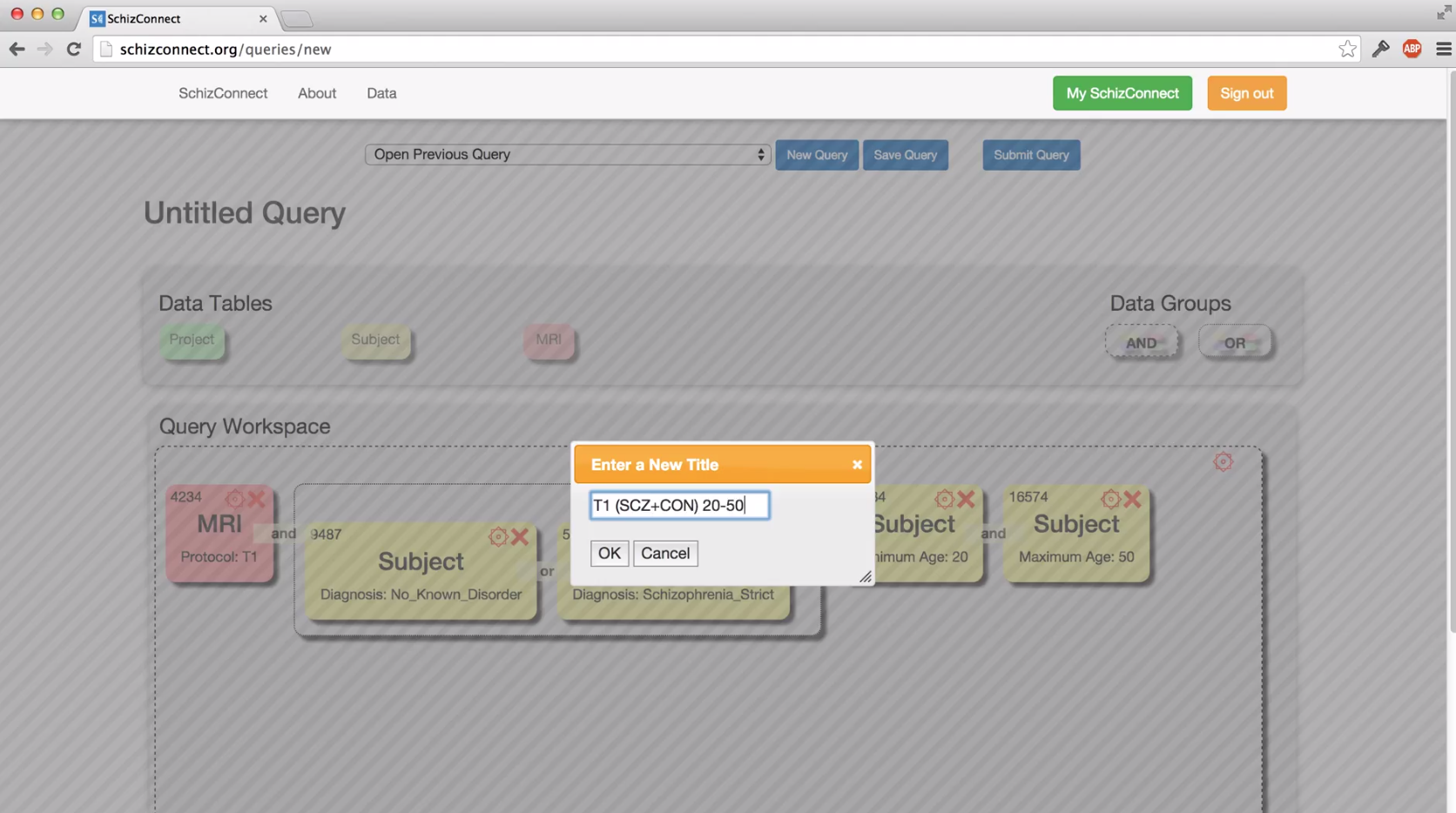
Would you explain more about how to use "AND" versus "OR" in my query?
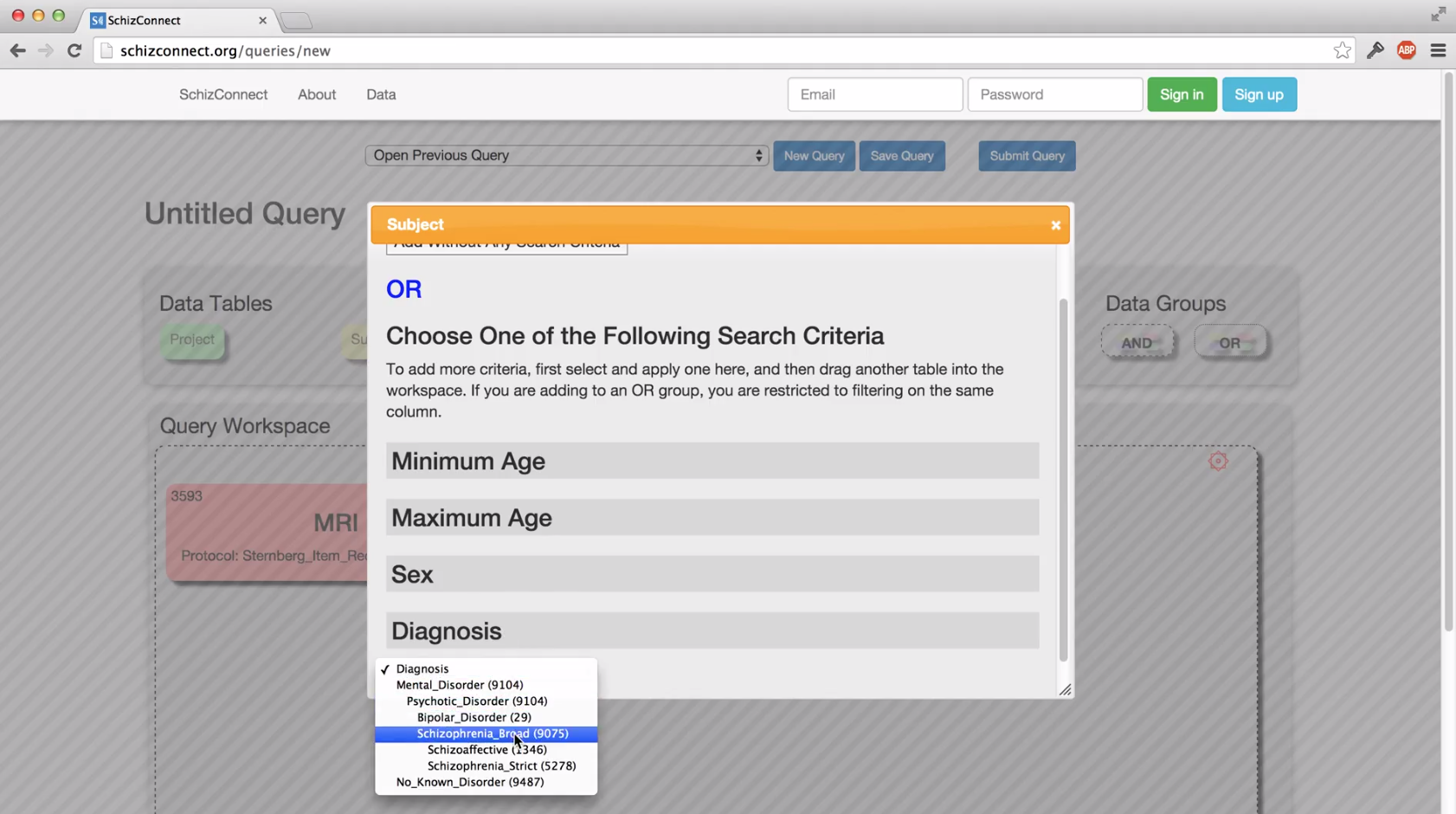
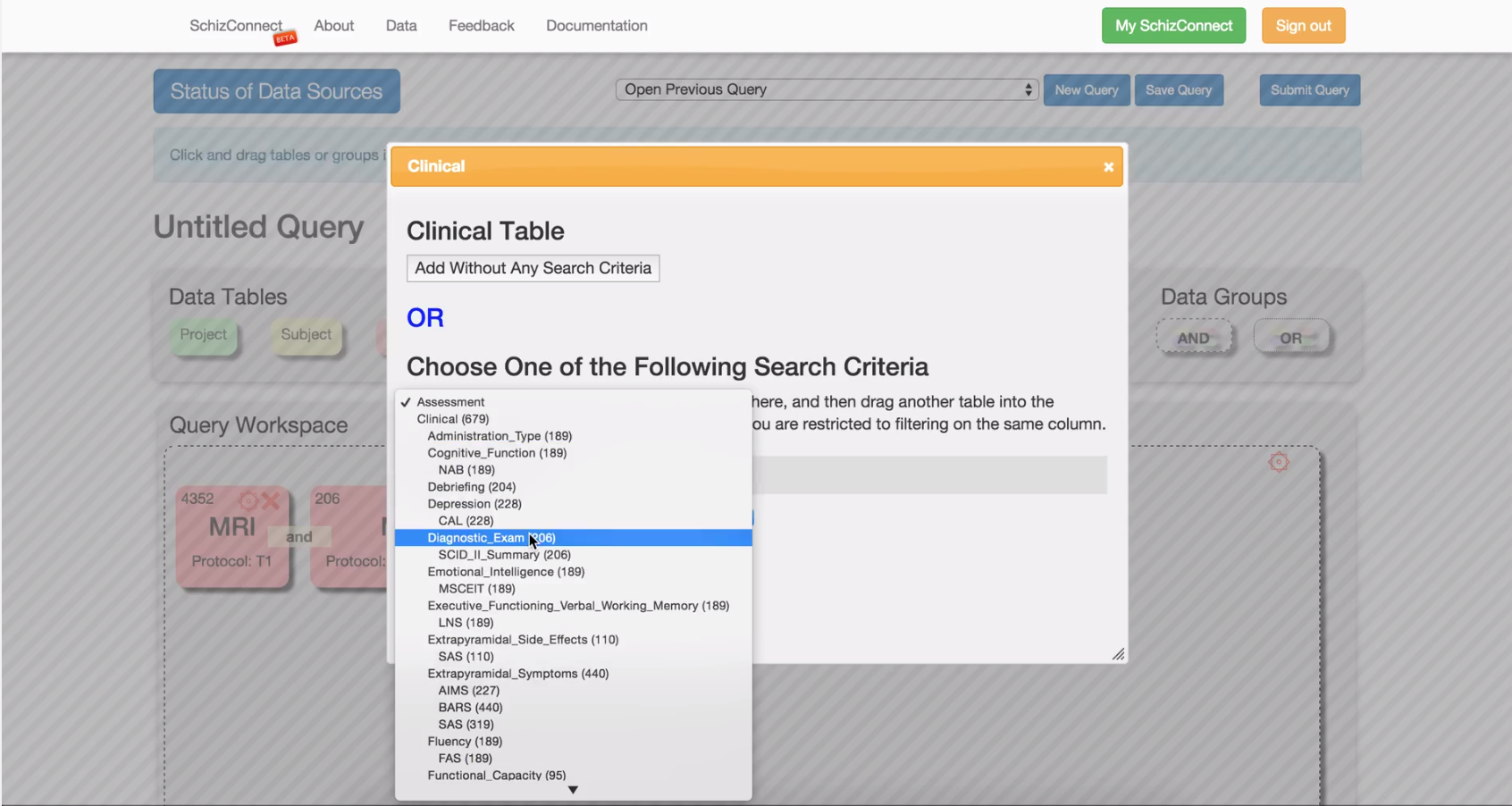
Can I query for subjects with certain clinical or cognitive assessment results?
If you are interested in clinical diagnosis, sex, or age, then yes: drag in the "Subject" table and filter as desired (or select "Add Without Any Search Criteria" to see this information for all subjects your query returns without any additional filtering).
For the clinical and cognitive assessments available from the "Clinical" or "Cognitive" tables, you can only filter on whether a given assessment exists for a subject, not the assessment results. Thus, when you see the option for "Any Extrapyramidal_Symptoms" (for example), this means "return subjects that have undergone any Extrapyramidal_Symptoms assessments," not "return subjects who exhibit extrapyramidal symptoms."
Where can I find demographic information for the subjects in my query?
You can get basic demographics (age, sex, diagnosis) for the subjects returned in a query by dragging the "Subjects" table into the query and selecting "Add Without Any Search Criteria".
Once you have initiated a downloaded for a query, you cannot modify it, but you can clone it, add the necessary table, and re-download. It is wise to wait until your original download completes before initiating the second download (so that the imaging data from the original query can be reused, as opposed to collected again from the remote sites). As long as you initiate your second download within 2 weeks of receiving the package from your original query, the downloaded imaging data from the original query will be reused and your results will be available very quickly. The cloned query's *metaData*.csv file will contain the age, sex, and diagnosis for the subjects.
If you are interested in other demographics information (handedness, SES, etc), you'll want to drag the "Clinical" table into the query and select Demographics>"Any Demographics". It's possible that some subjects will not have a demographics clinical assessment, so adding this restriction may narrow your results and require an entirely new package to be downloaded.
How do I query for longitudinal images or cross-sectional images?
To restrict your query to cross-sectional images only, drag the "MRI" table into your query and scroll down to select "Visit" > "Cross-sectional". This will return images from 1) subjects without follow-up imaging sessions and 2) baseline sessions for subjects with follow-ups.
To restrict your query to longitudinal images only (that is: images from subjects with a baseline and one or more follow-up sessions), drag the "MRI" table into your query and scroll down to select "Visit" > "Longitudinal (baseline and any follow-ups)". This will return all baseline and follow-up images for subjects with longitudinal imaging data.
Do you have an API?
There is a SchizConnect API for obtaining overall counts and metadata and assessment data for all subjects in SchizConnect; imaging files and any sort of filtered result can only be obtained by running a query through the website and submitting it for download.
To use the Schizconnect API, follow the instructions below:
- To get counts only, you do not need to be signed in. Simply run the command below:
curl http://schizconnect.org/api/tables/mri/listing
In place of mri, you may query the project, subject, mri, clinical, or cognitive table. - To get data, you must sign up for a SchizConnect account and sign the SchizConnect data use agreement. The project table result is available to all SchizConnect users who have signed the SchizConnect DUA. To see results for other tables, you must sign the project-specific data use agreements.
- The API validates against a token. Obtain your token by running the command below, replacing your_email@example.com and your_password with the email and password you use to login to SchizConnect:
curl -X POST -v -H 'Content-Type: application/json' http://schizconnect.org/api/auth/sign_in -d '{"email": "your_email@example.com", "password": "your_password"}' 2>&1 | egrep -i '(access-token|client|uid)' - This should return something like the below:
< access-token: my_access_token
< client: my_client_token
< uid: my_uid - Using the tokens obtained above, you may now query the API as follows:
curl -H 'access-token: my_access_token' -H 'client: my_client_token' -H 'uid: my_uid' http://schizconnect.org/api/tables/mri/listing
In place of mri, you may query the project, subject, mri, clinical, or cognitive table.
Can I use the API for downloading query results?
Yes! Follow the instructions in the API FAQ to obtain your credentials. This should return something like the below:
< access-token: my_access_token
< client: my_client_token
< uid: my_uid
Using the above token information, you may now download your package, replacing "http://schizconnect.org/downloadable_files/1" in the command below with the path to your download package file (you can obtain this by right-clicking on the link in your results email and selecting "Copy Link"). You may also wish to replace the output file name ("package_file_1.7z.001", below) with the name of the file in the results email.
curl -H 'access-token: my_access_token' -H 'client: my_client_token' -H 'uid: my_uid' http://schizconnect.org/downloadable_files/1 -o package_file_1.7z.001
Are you planning to add any sort of processing pipelines to SchizConnect?
Yes! We are planning to incorporate some sort of workflow processing into SchizConnect. You may wish to join our mailing list to stay up-to-date on our progress.
What is the difference between the "schizophrenia broad" and "schizophrenia strict" diagnosis categories?
The schizophrenia broad category includes those diagnosed with schizophrenia according to the DSM IV (the schizophrenia strict category) and also those with schizoaffective disorder (the schizoaffective category).
Are all SchizConnect downloads in BIDS format?
Not quite. We are still working to convert some of the fMRI behavioral data into BIDS format (until we do, you will find it in raw edat/txt files alongside the images or in log files in a "beh" directory). The images themselves should all be in BIDS format. We thought having all images in a consistent directory and file structure was useful enough to warrant early deployment with this behavioral data caveat, rather than waiting on the behavioral data, which requires expert input for BIDS conversion.
Are there any subjects that have been scanned for multiple projects?
Where can I find functional Slice Ordering information?
For all projects except fBIRN, you should use the BIDS json sidecar as described in this thread. For fBIRN, the information is contained in the *.ImageWrapper.xml file.
What if my question isn't answered here?
Tutorials
Data Use Agreements
Recipient (identified below) is requesting access to SchizConnect for the purpose of scientific investigation, teaching or the planning of clinical research studies. Prior to accessing the SchizConnect data ("Data"), Recipient must agree to the following terms and conditions regarding its use:
- Recipient will receive access to de-identified Data which are to be used at Recipient’s own risk. The Data are provided "as is" and no warranties are provided, express or implied, regarding the Data, its merchantability or fitness for a particular purpose, its use by Recipient or any product or process based thereon.
- Recipient agrees that the Data should not be redistributed to third parties unless such third parties have agreed in writing to the terms and conditions contained herein prior to transfer. Recipient agrees that appropriate safeguards are in place to ensure that the Data is not used or disclosed in a manner inconsistent with the terms of this agreement. Recipient agrees to report any unauthorized use of the Data within fifteen (15) days of Recipient becoming aware of such unauthorized use or disclosure.
- Recipient will make no attempt to
determine the identity of any individual who contributed to these Data, except
as permitted by law.
- Recipient agrees to obtain any
necessary approvals or permissions which may be required by its institutional
review board or human subjects protections office regarding the use of
SchizConnect Data.
- Recipient will cite SchizConnect as the source of the Data, and the SchizConnect funding source (1U01 MH097435) as scientifically appropriate or customary in any abstract, publication or public disclosure reporting Recipient’s use of the Data in order to accurately acknowledge the contributions of SchizConnect personnel. Depending upon the length and focus of the article, it may be appropriate to include more or less than the example below, however, inclusion of some variation of the language shown below is mandatory:
- "Data used in preparation of this article were obtained from the SchizConnect database (http://schizconnect.org) As such, the investigators within SchizConnect contributed to the design and implementation of SchizConnect and/or provided data but did not participate in analysis or writing of this report."
- Recipient will acknowledge funding by SchizConnect in the support acknowledgement section of the manuscript using language similar to the following:
- "Data collection and sharing for this project was funded by NIMH cooperative agreement 1U01 MH097435."
- It is the policy of SchizConnect to make Data available to investigators as quickly as possible. However, data analysis for this project is expected to evolve as methods and procedures develop over time. Therefore, Recipient acknowledges that any Data downloaded by Recipient may be preliminary and that results may change as new methods of analysis are implemented. Recipient should ensure it is using the most recent Data available, prior to publication.
- Recipient understands that failure to abide by these terms and conditions will result in termination of its privileges to access SchizConnect Data.
Revised October 2014
Acknowledgement: Data was downloaded from the COllaborative Informatics and Neuroimaging Suite Data Exchange tool (COINS;http://coins.mrn.org/dx) and data collection was funded by NIMH R01MH084898-01A1,
“Brain Glutamate and Outcome in Schizophrenia”, PI: J. Bustillo Bustillo JR, et al, Glutamatergic and neuronal dysfunction in gray and white matter: a spectroscopic imaging study in a large schizophrenia sample Schizophr Bull. 2016 Aug 22. PubMed PMID: 27550776.
Please agree to include the acknowledgment and cite the article below in any manuscript in which the COBRE data is used.
Acknowledgement: Data was downloaded from the COllaborative Informatics and Neuroimaging Suite Data Exchange tool (COINS; http://coins.mrn.org/dx) and data collection was performed at the Mind Research Network, and funded by a Center of Biomedical Research Excellence (COBRE) grant 5P20RR021938/P20GM103472 from the NIH to Dr. Vince Calhoun.
C. Aine, H. J. Bockholt, J. Bustillo, J. Canive, A. Caprihan, C. Gasparovic, F. M. Hanlon, J. Houck, R. Jung, J. Lauriello, J. Liu, A. Mayer, N. Perrone-Bizzozero, S. Posse, J. Stephen, J. Turner, V. Clark, and V. D. Calhoun, "Multimodal Imaging in Schizophrenia: Description and Dissemination," NeuroInformatics, in press. https://www.ncbi.nlm.nih.gov/pubmed/28812221
FBIRN Data Use Agreement
I request access to data collected by the Mind Clinical Imaging Consortium (MCIC) for the purpose of scientific investigation, teaching or the planning of clinical research studies and agree to the following terms:
1. I will receive access to de-identified data and will not attempt to establish the identity of, or attempt to contact any of the MCIC participants.2. I will not further disclose these data beyond the uses outlined in this agreement and my data use application.
3. I will require anyone on my team who utilizes these data, or anyone with whom I share these data to comply with this data use agreement.
4. I will comply with any rules and regulations imposed by my institution and its institutional review board in requesting these data.
5. I will ensure that Investigators who utilize MCIC data use appropriate administrative, physical and technical safeguards to prevent use or disclosure of the data other than as provided for by this Agreement.
6. I will report any use or disclosure of the data not provided for by this Agreement of which I become aware within 15 days of becoming aware of such use or disclosure. Reports/disclosures should be sent via email to Jody Roberts, Director of Research and Clinical Operations at MRN, jroberts@mrn.org
If I publish abstracts or manuscripts using data from MCIC, I agree to the following:
7. I will cite MCIC as the source of data and the DOE funding source in the publication as space allows. Ex: "The imaging data and demographic information was collected and shared by [University of Iowa, University of Minnesota, University of New Mexico, Massachusetts General Hospital] the Mind Research Network supported by the Department of Energy under Award Number DE-FG02-08ER64581."
8. I will cite Gollub, RL, et al “The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia” Neuroinformatics, 2013 in all reports based on these data, as appropriate.
9. I will include language similar to the following in the methods section of my manuscripts in order to accurately acknowledge data gathering by the MCIC investigators. Depending upon the length and focus of the article, it may be appropriate to include more or less than the example below, however, inclusion of some variation of the language shown below is mandatory. "Data used in the preparation of this work were obtained from the Mind Clinical Imaging Consortium database through the Mind Research Network (www.mrn.org). The MCIC project was supported by the Department of Energy under Award Number DE-FG02-08ER64581. MCIC is the result of efforts of co-investigators from University of Iowa, University of Minnesota, University of New Mexico, Massachusetts General Hospital.
I understand that failure to abide by these guidelines will result in termination of my privileges to access MCIC data.
Recipient (identified below) is requesting access to the Neuromorphometry by Computer Algorithm Chicago (NMorphCH) dataset for the purpose of scientific investigation, teaching or the planning of clinical research studies. Prior to accessing the NMorphCH data ("Data"), Recipient must agree to the following terms and conditions regarding its use:
- Recipient will receive access to de-identified Data which are to be used at Recipient’s own risk. The Data are provided "as is" and Northwestern University makes no warranties, express or implied, regarding the Data, its merchantability or fitness for a particular purpose, its use by Recipient or any product or process based thereon.
- Recipient will make no attempt to determine the identity of any individual who contributed to these Data.
- Recipient will cite NMorphCH as the source of Data, and the NMorphCH funding source (R01 MH056584) in the abstract as space allows.
- Recipient will include appropriate acknowledgement of the source of the Data in any manuscripts or publications reporting its use in order to accurately acknowledge the contributions of NMorphCH personnel. Depending upon the length and focus of the article, it may be appropriate to include more or less than the example below, however, inclusion of some variation of the language shown below is mandatory:
- "Data used in preparation of this article were obtained from the Neuromorphometry by Computer Algorithm Chicago (NMorphCH) dataset (http://nunda.northwestern.edu/nunda/data/projects/NMorphCH) As such, the investigators within NMorphCH contributed to the design and implementation of NMorphCH and/or provided data but did not participate in analysis or writing of this report."
- Recipient will acknowledge funding by NMorphCH in the support acknowledgement section of the manuscript using language similar to the following:
- "Data collection and sharing for this project was funded by NIMH grant R01 MH056584."
- It is the policy of NMorphCH to make Data available to investigators as quickly as possible. However, data analysis for this project is expected to evolve as methods and procedures develop over time. Therefore, Recipient acknowledges that any Data downloaded by Recipient may be preliminary and that results may change as new methods of analysis are implemented. Recipient should ensure it is using the most recent Data available, prior to publication.
- Recipient agrees that the Data should not be redistributed to third parties unless such third parties have agreed in writing to the terms and conditions contained herein prior to transfer.
- Recipient understands that failure to abide by these terms and conditions will result in termination of its privileges to access NMorphCH Data.
Revised September 2016
Recipient (identified below) is requesting access to Northwestern University Schizophrenia Data and Software Tool for the purpose of scientific investigation, teaching or the planning of clinical research studies. Prior to accessing the NUSDAST data ("Data"), Recipient must agree to the following terms and conditions regarding its use:
- Recipient acknowledges that it may also request data access on XNAT Central.
- Recipient will receive access to de-identified Data which are to be used at Recipient’s own risk. The Data are provided "as is" and Northwestern University makes no warranties, express or implied, regarding the Data, its merchantability or fitness for a particular purpose, its use by Recipient or any product or process based thereon.
- Recipient will make no attempt to determine the identity of any individual who contributed to these Data.
- Recipient will cite NUSDAST as the source of Data, and the NUSDAST funding source (1R01 MH084803) in the abstract as space allows.
- Recipient will include appropriate acknowledgement of the source of the Data in any manuscripts or publications reporting its use in order to accurately acknowledge the contributions of NUSDAST personnel. Depending upon the length and focus of the article, it may be appropriate to include more or less than the example below, however, inclusion of some variation of the language shown below is mandatory:
- "Data used in preparation of this article were obtained from the NU Schizophrenia Data and Software Tool (NUSDAST) database (http://central.xnat.org/REST/projects/NUDataSharing) As such, the investigators within NUSDAST contributed to the design and implementation of NUSDAST and/or provided data but did not participate in analysis or writing of this report."
- Recipient will acknowledge funding by NUSDAST in the support acknowledgement section of the manuscript using language similar to the following:
- "Data collection and sharing for this project was funded by NIMH grant 1R01 MH084803."
- Recipient agrees to provide a bibliographic citation of the final published presentation or article for inclusion in the NUSDAST literature archive. Recipient will send this information to leiwang1@northwestern.edu
- It is the policy of NUSDAST to make Data available to investigators as quickly as possible. However, data analysis for this project is expected to evolve as methods and procedures develop over time. Therefore, Recipient acknowledges that any Data downloaded by Recipient may be preliminary and that results may change as new methods of analysis are implemented. Recipient should ensure it is using the most recent Data available, prior to publication.
- Recipient agrees that the Data should not be redistributed to third parties unless such third parties have agreed in writing to the terms and conditions contained herein prior to transfer.
- Recipient understands that failure to abide by these terms and conditions will result in termination of its privileges to access NUSDAST Data.
Revised October 2014
Recipient (identified below) is requesting access to the REWARD dataset for the purpose of scientific investigation, teaching or the planning of clinical research studies. Prior to accessing the REWARD data ("Data"), Recipient must agree to the following terms and conditions regarding its use:
- Recipient will receive access to de-identified Data which are to be used at Recipient’s own risk. The Data are provided "as is" and Washington University in St. Louis makes no warranties, express or implied, regarding the Data, its merchantability or fitness for a particular purpose, its use by Recipient or any product or process based thereon.
- Recipient will make no attempt to determine the identity of any individual who contributed to these Data.
- Recipient will cite REWARD as the source of Data, and the REWARD funding source (NIMH MH066031) in abstracts for submission and the methods sections of papers.
- Recipient will include appropriate
acknowledgement of the source of the Data in any manuscripts or
publications reporting its use in order to accurately acknowledge the
contributions of REWARD personnel. Depending upon the length and focus of
the article, it may be appropriate to include more or less than the
example below, however, inclusion of some variation of the language shown
below is mandatory:
- "Data used in preparation of this article were obtained from the REWARD dataset. As such, the investigators within REWARD contributed to the design and implementation of REWARD and/or provided data but did not participate in analysis or writing of this report."
- Recipient will acknowledge funding
by REWARD in the support acknowledgement section of the manuscript using
language similar to the following:
- "Data collection and sharing for this project was funded by NIMH grant MH066031."
- It is the policy of REWARD to make Data available to investigators as quickly as possible. However, data analysis for this project is expected to evolve as methods and procedures develop over time. Therefore, Recipient acknowledges that any Data downloaded by Recipient may be preliminary and that results may change as new methods of analysis are implemented. Recipient should ensure it is using the most recent Data available, prior to publication.
- Recipient agrees that the Data should not be redistributed to third parties unless such third parties have agreed in writing to the terms and conditions contained herein prior to transfer.
- Recipient understands that failure to abide by these terms and conditions will result in termination of its privileges to access REWARD Data.
Revised November 2017
Data Models With Links to Ontology
MRI Table: SchizConnect Imaging Protocol
Subject Table: SchizConnect Subject Model
Imaging Data Information
FBIRN_Data_Hierarchy_Description
Data Dictionaries
BrainGluSchi: BrainGluSchi_Data_Dictionary
COBRE: COBRE_Data_Dictionary
fBIRNPhaseII__0010: HID_PhaseII_Assessments_DataDictionary_v2
MCICShare: MCIC_Data_Dictionary
Publications
SchizConnect Publications
-
Ambite, J., Tallis, M., Alpert, K., Keator, D., King, M., Landis, D., Konstantinidis, G., Calhoun, V., Potkin, S., Turner, J., Wang, L., 2015. SchizConnect: Virtual Data Integration in Neuroimaging. In: Ashish, N., Ambite, J.-L. (Eds.), Data Integration in the Life Sciences. Springer International Publishing, pp. 37-51.
-
Wang, L., Alpert, K.I., Calhoun, V.D., Cobia, D.J., Keator, D.B., King, M.D., Kogan, A., Landis, D., Tallis, M., Turner, M.D., Potkin, S.G., Turner, J.A., Ambite, J.L., 2016. SchizConnect: Mediating neuroimaging databases on schizophrenia and related disorders for large-scale integration. Neuroimage 124, 1155-1167.
-
Turner, J., Pasquerello, D., Turner, M., Keator, D., Alpert, K., King, M., Landis, D., Calhoun, V., Potkin, S., Tallis, M., Ambite, J., Wang, L., 2015. Terminology Development Towards Harmonizing Multiple Clinical Neuroimaging Research Repositories. In: Ashish, N., Ambite, J.-L. (Eds.), Data Integration in the Life Sciences. Springer International Publishing, pp. 104-117.
REWARD Publications
-
Barch, D. M., Treadway, M., & #Schoen, N. (2014). Effort, anhedonia, and function in schizophrenia: Reduced effort allocation predicts amotivation and functional impairment. Journal of Abnormal Psychology, 123, 387-397. PMCID: PMC4048870
-
Becerril, K. E. & Barch, D. M. (2011). Influence of emotional processing on working memory in schizophrenia. Schizophrenia Bulletin, 37, 1027-1038. PMCID: PMC3160211
-
Becerril, K. E. & Barch, D. M. (2013). Beyond the anterior cingulate cortex: Conflict and error processing in an extended cingulo-opercular network in schizophrenia. Neuroimage: Clinical, 3, 470-480. PMCID: PMC3830057
-
Becerril, K. E., Repovs, G., & Barch, D. M. (2011). Error processing network dynamics in schizophrenia. Neuroimage, 54, 1495-1505. PMCID: PMC2997133
-
Chung, Y. S. & Barch, D. M. (2015). Anhedonia is associated with reduced incentive cue related activation in the basal ganglia. Cognitive, Affective and Behavioral Neuroscience, 15(4), 749-767. PMCID: PMC5141384
-
Chung, Y. S. & Barch, D. M. (2016). Frontal-striatal dysfunction during reward processing; Relationships to amotivation in schizophrenia. Journal of Abnormal Psychology, 125, 453-469. PMCID: PMC5132953
-
Culbreth, A. J., Gold, J. M., Cools, R., & Barch, D. M. (2016). Impaired activation in cognitive control regions predicts performance during reversal learning in schizophrenia. Schizophrenia Bulletin, 42, 484-493. PMCID: PMC4753588
-
Culbreth AJ, Westbrook JA, Xu Z, Barch DM, Waltz JA. (2016). Intact Ventral Striatal Prediction Error Signaling in Medicated Schizophrenia Patients. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. September 16; 1(5): 474- 483. PMC Journal – In Process, NIHMSID: NIHMS809970
-
Dowd, E., & Barch, D. M. (2012). Reward anticipation and receipt during Pavlovian conditioning in schizophrenia: Relationship to anhedonia. PLOS One, 7, e35622. PMCID: PMC3344823
-
Dowd, E. C. & Barch, D. M. (2010). Subjective emotional experience in schizophrenia: Neural and behavioral markers. Biological Psychiatry, 15, 902-911. PMCID: PMC3113677
-
Dowd, E. C., Frank, M., Collins, A., & Barch, D. M. (2016). Probabilistic reinforcement learning in schizophrenia: Relationships to anhedonia and avolition. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 1, 460-473. PMCID: PMC509850
-
Lerman-Sinkoff, D. B. & Barch, D. M. (2016). Network community structure alterations in adult schizophrenia: identification and localization of alterations. Neuroimage: Clinical, 10, 97-106. PMCID: PMC4683428
-
Mann, C. L., Footer, O., Chung, Y. S., Driscoll, L. L., & Barch, D. M. (2013). Spared and impaired aspects of motived cognitive control in schizophrenia. Journal of Abnormal Psychology, 122, 745-755. PMCID: PMC3863584
BrainGluSchi Publications
-
Bustillo JR, et al, Glutamatergic and neuronal dysfunction in gray and white matter: a spectroscopic imaging study in a large schizophrenia sample Schizophr Bull. 2016 Aug 22. PubMed PMID: 27550776.
COBRE Publications
-
C. Aine, H. J. Bockholt, J. Bustillo, J. Canive, A. Caprihan, C. Gasparovic, F. M. Hanlon, J. Houck, R. Jung, J. Lauriello, J. Liu, A. Mayer, N. Perrone-Bizzozero, S. Posse, J. Stephen, J. Turner, V. Clark, and V. D. Calhoun, "Multimodal Imaging in Schizophrenia: Description and Dissemination," NeuroInformatics, in press.
-
M. Çetin, F. Christensen, C. Abbott, J. Stephen, A. Mayer, J. Cañive, J. Bustillo, G. Pearlson, and V. D. Calhoun, "Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia," NeuroImage, vol. 97, pp. 117-126, 2014.
fBIRNPhaseII__0010 Publications
-
Gadde, S., Aucoin, N., Grethe, J. S., Keator, D. B., Marcus, D. S., Pieper, S., FBIRN, MBIRN, BIRN-CC. (2012). XCEDE: an extensible schema for biomedical data. Neuroinformatics, 10(1), 19–32. http://doi.org/10.1007/s12021-011-9119-9
-
Glover, G. H., Mueller, B. A., Turner, J. A., van Erp, T. G. M., Liu, T. T., Greve, D. N., et al. (2012). Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. Journal of Magnetic Resonance Imaging, 36(1), 39–54. http://doi.org/10.1002/jmri.23572
-
Ozyurt, I. B., Keator, D. B., Wei, D., Fennema-Notestine, C., Pease, K. R., Bockholt, J., & Grethe, J. S. (2010). Federated Web-accessible Clinical Data Management within an Extensible NeuroImaging Database. Neuroinformatics, 8(4), 231–249. http://doi.org/10.1007/s12021-010-9078-6
MCICShare Publications
-
Gollub, R. L., Shoemaker, J. M., King, M. D., White, T., Ehrlich, S., Sponheim, S. R., … Andreasen, N. C. (2013). The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics, 11(3), 367–388. doi:10.1007/s12021-013-9184-3
NUSDAST Publications
-
Kogan, A., Alpert, K., Ambite, J. L., Marcus, D. S., & Wang, L. (2016). Northwestern University schizophrenia data sharing for SchizConnect: A longitudinal dataset for large-scale integration. NeuroImage, 124, Part B, 1196–1201. http://doi.org/10.1016/j.neuroimage.2015.06.030
-
Wang, Lei, Alexander Kogan, Derin Cobia, Kathryn Alpert, Anthony Kolasny, Michael I. Miller, Daniel Marcus. Northwestern University Schizophrenia Data and Software Tool (NUSDAST). Frontiers in Neuroinformatics 2013; 7:25
Documentation by Project
BrainGluSchi
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
-
Bustillo JR, et al, Glutamatergic and neuronal dysfunction in gray and white matter: a spectroscopic imaging study in a large schizophrenia sample Schizophr Bull. 2016 Aug 22. PubMed PMID: 27550776.
COBRE
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
-
C. Aine, H. J. Bockholt, J. Bustillo, J. Canive, A. Caprihan, C. Gasparovic, F. M. Hanlon, J. Houck, R. Jung, J. Lauriello, J. Liu, A. Mayer, N. Perrone-Bizzozero, S. Posse, J. Stephen, J. Turner, V. Clark, and V. D. Calhoun, "Multimodal Imaging in Schizophrenia: Description and Dissemination," NeuroInformatics, in press.
-
M. Çetin, F. Christensen, C. Abbott, J. Stephen, A. Mayer, J. Cañive, J. Bustillo, G. Pearlson, and V. D. Calhoun, "Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia," NeuroImage, vol. 97, pp. 117-126, 2014.
fBIRNPhaseII__0010
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
-
Gadde, S., Aucoin, N., Grethe, J. S., Keator, D. B., Marcus, D. S., Pieper, S., FBIRN, MBIRN, BIRN-CC. (2012). XCEDE: an extensible schema for biomedical data. Neuroinformatics, 10(1), 19–32. http://doi.org/10.1007/s12021-011-9119-9
-
Glover, G. H., Mueller, B. A., Turner, J. A., van Erp, T. G. M., Liu, T. T., Greve, D. N., et al. (2012). Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. Journal of Magnetic Resonance Imaging, 36(1), 39–54. http://doi.org/10.1002/jmri.23572
-
Ozyurt, I. B., Keator, D. B., Wei, D., Fennema-Notestine, C., Pease, K. R., Bockholt, J., & Grethe, J. S. (2010). Federated Web-accessible Clinical Data Management within an Extensible NeuroImaging Database. Neuroinformatics, 8(4), 231–249. http://doi.org/10.1007/s12021-010-9078-6
MCICShare
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
-
Gollub, R. L., Shoemaker, J. M., King, M. D., White, T., Ehrlich, S., Sponheim, S. R., … Andreasen, N. C. (2013). The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics, 11(3), 367–388. doi:10.1007/s12021-013-9184-3
NMorphCH
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
NUSDAST
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
-
Kogan, A., Alpert, K., Ambite, J. L., Marcus, D. S., & Wang, L. (2016). Northwestern University schizophrenia data sharing for SchizConnect: A longitudinal dataset for large-scale integration. NeuroImage, 124, Part B, 1196–1201. http://doi.org/10.1016/j.neuroimage.2015.06.030
-
Wang, Lei, Alexander Kogan, Derin Cobia, Kathryn Alpert, Anthony Kolasny, Michael I. Miller, Daniel Marcus. Northwestern University Schizophrenia Data and Software Tool (NUSDAST). Frontiers in Neuroinformatics 2013; 7:25
REWARD
Data Use Agreement
Imaging Data Information
Data Dictionary
Publications
-
Barch, D. M., Treadway, M., & #Schoen, N. (2014). Effort, anhedonia, and function in schizophrenia: Reduced effort allocation predicts amotivation and functional impairment. Journal of Abnormal Psychology, 123, 387-397. PMCID: PMC4048870
-
Becerril, K. E. & Barch, D. M. (2011). Influence of emotional processing on working memory in schizophrenia. Schizophrenia Bulletin, 37, 1027-1038. PMCID: PMC3160211
-
Becerril, K. E. & Barch, D. M. (2013). Beyond the anterior cingulate cortex: Conflict and error processing in an extended cingulo-opercular network in schizophrenia. Neuroimage: Clinical, 3, 470-480. PMCID: PMC3830057
-
Becerril, K. E., Repovs, G., & Barch, D. M. (2011). Error processing network dynamics in schizophrenia. Neuroimage, 54, 1495-1505. PMCID: PMC2997133
-
Chung, Y. S. & Barch, D. M. (2015). Anhedonia is associated with reduced incentive cue related activation in the basal ganglia. Cognitive, Affective and Behavioral Neuroscience, 15(4), 749-767. PMCID: PMC5141384
-
Chung, Y. S. & Barch, D. M. (2016). Frontal-striatal dysfunction during reward processing; Relationships to amotivation in schizophrenia. Journal of Abnormal Psychology, 125, 453-469. PMCID: PMC5132953
-
Culbreth, A. J., Gold, J. M., Cools, R., & Barch, D. M. (2016). Impaired activation in cognitive control regions predicts performance during reversal learning in schizophrenia. Schizophrenia Bulletin, 42, 484-493. PMCID: PMC4753588
-
Culbreth AJ, Westbrook JA, Xu Z, Barch DM, Waltz JA. (2016). Intact Ventral Striatal Prediction Error Signaling in Medicated Schizophrenia Patients. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. September 16; 1(5): 474- 483. PMC Journal – In Process, NIHMSID: NIHMS809970
-
Dowd, E., & Barch, D. M. (2012). Reward anticipation and receipt during Pavlovian conditioning in schizophrenia: Relationship to anhedonia. PLOS One, 7, e35622. PMCID: PMC3344823
-
Dowd, E. C. & Barch, D. M. (2010). Subjective emotional experience in schizophrenia: Neural and behavioral markers. Biological Psychiatry, 15, 902-911. PMCID: PMC3113677
-
Dowd, E. C., Frank, M., Collins, A., & Barch, D. M. (2016). Probabilistic reinforcement learning in schizophrenia: Relationships to anhedonia and avolition. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 1, 460-473. PMCID: PMC509850
-
Lerman-Sinkoff, D. B. & Barch, D. M. (2016). Network community structure alterations in adult schizophrenia: identification and localization of alterations. Neuroimage: Clinical, 10, 97-106. PMCID: PMC4683428
-
Mann, C. L., Footer, O., Chung, Y. S., Driscoll, L. L., & Barch, D. M. (2013). Spared and impaired aspects of motived cognitive control in schizophrenia. Journal of Abnormal Psychology, 122, 745-755. PMCID: PMC3863584